
Welcome to our SDG Editorial Board Members blog collection. We are hearing from the Editorial Board Members of the BMC Series journals whose work aligns with achieving the Sustainable Development Goals. Here you can find other posts in this collection, grouped with the tag ‘SDG editorial board members‘.
My work relates to the WHO Sustainable Development Goal SDG3 and focuses on ensuring healthy lives and promoting well-being for all. I have experience in microbial genomics and computational approaches. My research looks at the use of genomic methods for pathogen detection, characterisation as well as understanding transmission dynamics and acquisition of antimicrobial resistance at hospitals and community settings. I am proud to collaborate in achieving SDG3 by working with National Public Health Institutes (NPHI) in Africa facilitating access to sustainable DNA sequencing technologies for rapid and timely response to common infectious disease threats, including drug resistance. Recently, my group has developed an automated & highly scalable assembly, annotation, and higher-level analyses microbial computational pipeline for genomic data called rMAP: Rapid microbial analysis, profiling pipeline, and visualisation (https://bit.ly/36g809L).
Some of the challenges I have encountered during my research include limited access to DNA sequencing technologies, competitive funding, or high-performance computing (HPC) bioinformatic environments to support large-scale data analyses. However, I have established research collaborations and networks to increase funding opportunities and access to DNA sequencing technologies and HPC. For example, in 2019, the U.S. National Institutes of Health launched the second African Center of Excellence in Bioinformatics (ACE) at the Infectious Diseases Institute in Makerere University’s College (https://ace.idi.co.ug/). This center serves as a regional bioinformatics HPC resource for training and collaborative research for institutions in Africa.

Tools that read and interpret gene variations to enable customized medical care during HIV testing, treatment & management
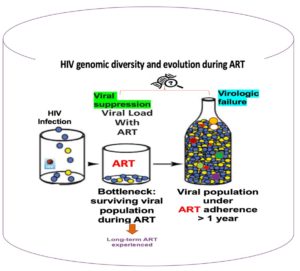
HIV infection results from the expansion of a single or limited number of viral variants (genetically different copies of the virus). The differences in HIV virus come into existence after stable changes occur in its genes. This can happen during the exchange of genetic material between different HIV viruses in the body or after certain viral gene combinations become more prevalent than others due to exposure to antiretroviral therapy (ART) regimens. But also, sometimes viruses do not naturally develop smoothly from one stage to the next in a logical way. ART, the drugs used to treat HIV/AIDS, continue to save the lives of tens of millions of people living with HIV/AIDS (PLWHA). At the end of 2020, 27.5 million people, out of an estimated 37.7 million PLWHA, were receiving ART globally. This increased use of ART has been accompanied by the emergence of HIV drug resistance (HIVDR) – the levels of which have steadily increased in recent years. HIVDR is caused by changes in HIV genes that affect the ability of drugs to stop the multiplication of the virus. All ART drugs are at risk of becoming partly or fully inactive due to the emergence of drug-resistant virus strains. HIVDR leads to increased numbers of HIV infections and HIV-associated illnesses and deaths. The World Health Organization (WHO) report revealed that in 2020, 64% of countries with a high burden of HIV infections had growing HIVDR and recommended national action plans to prevent, monitor and respond to HIVDR. HIV virological failure (VF) signals treatment effectiveness and occurs when ART fails to suppress and sustain one’s viral load to <200 viral copies per ml and is normally associated with HIVDR. It is estimated that 24% of HIV patients in Sub-Saharan Africa have experienced VF. HIVDR and VF remain barriers to reaching the Joint United Nations Programme on HIV/AIDS (UNAIDS) fast-track goal of ending AIDS by 2030.
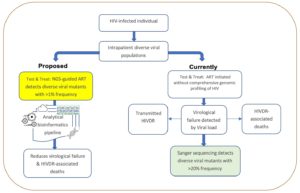
Sanger sequencing (method of gene sequencing involving electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides) is the gold standard for HIVDR testing but is limited by non-detection of resistance-associated variants with prevalence <20% yet these cause VF. Next-generation sequencing or NGS (multiplies millions of copies of all different genes in the virus in a massively paralleled way) detects different HIV variants and has been implemented largely in high-income countries. However, there is very limited application of NGS-based HIVDR platforms in Sub-Saharan Africa which has nearly 70% of global HIV infections.
Why HIV NGS?
For HIV-infected individuals, comprehensive characterization of the different viral variants during initial testing before-ART initiation or during the treatment course improves their chances of survival by ensuring customized treatment while minimizing VF and HIVDR. Significant progress in HIV infection control has been realized in Sub-Saharan Africa over the last two decades largely due to increased access to ART. Advances such as second (Illumina, Roche 454, and Ion Torrent) or third-generation (PacBio and Nanopore) DNA sequencing technologies, that can accurately profile viral variants at any stage of HIV infection, are key for effective disease management, preventing VF, and HIVDR. Urgent identification of viral genetic markers that predict increased risk of VF or HIVDR in different ART-based regimens including newer ones like Dolutegravir and injectable long-acting Cabotegravir/Rilpivirine (CAB/RPV) is critical. This involves analysis of clinical alongside viral load data and viral gene variants using bioinformatics tools. The DNA sequencing technologies and advances in analytical approaches are poised to steer an era of personalized HIV management due to their ability to offer accurate rapid detection and drug resistance profiling close to point-of-care testing (also known as bedside testing).
Virological failure and drug resistance
Non-adherence to ART as well as poor monitoring of viral load during treatment are challenges that continue to affect HIV control programs in Africa leading to the emergence of HIVDR, virological failure, increased HIVDR transmissions, morbidity, and mortality. The WHO recommends global genomic surveillance to monitor dynamics of HIV diversity and emergence of HIVDR. Countries with high infection burden must build robust national HIV genomic surveillance programs to ensure improved management of PLWHA if they are to eradicate HIV/AIDS. ART also referred to as lifetime treatment has transformed HIV infection from a gradual, normally deadly illness to a prolonged disease that endures for many years while subjecting infected individuals to different ART treatments over time. Regular monitoring of viral load is currently used for the assessment of ART response in HIV management. The drugs cause changes in the genes of the virus. These viral gene changes can offer critical insights for understanding mechanisms of drug resistance, effects of therapeutics on the virus, and guide effective vaccine development.
Conclusion
Considering that many African countries have implemented the universal HIV test and treat strategy (which involves providing lifelong ART to PLWHA irrespective of CD4 or WHO HIV clinical staging), national HIV control programs should utilize NGS in routine HIV testing, treatment, and surveillance. However, this also requires appropriate policy changes, substantial investment in installing NGS platforms and bioinformatics capacity building for infectious diseases control programs.

Comments