
A new study by Dr. Nickolas Marra and colleagues on the transcriptomic analysis of elasmobranches (a subclass of cartilaginous fish such as sharks) and teleosts (a class of bony, ray-finned fishes) has revealed fascinating new information on evolutionary immunology and its impact on cancer development.
The researchers focus for this study was on seven particular species of fishes: four species of elasmobranchs (the white shark, shortfin mako, great hammerhead, and yellow stingray) and three species of teleosts (the swordfish, hogfish, and ocean surgeonfish), all of whom were not arbitrarily chosen. While the genomes of these organisms have not yet been extensively studied, they comprise a category of the oldest known vertebrates with a primitive adaptive immune system, efficient wound healing, and the development of regional endothermy in several species. This primitive immune system shares the same B and T cell genes which comprise the basis of our own adaptive immune system, and offers a glimpse into how this system has developed in a challenging environment.

The seven species that were studied are also known for having a low propensity for developing cancer (elasmobranchs are suspected to have the lowest incidence of malignant neoplasia of any vertebrate group) and illness associated with infection (based on several case studies), despite being known as the “garbage cans of the sea,” eating carcinogenic material and having flesh wounds exposed to bacteria filled ocean water. In theory this environment should increase their odds for developing tumors, but the opposite has shown to be true. The reason for this low disease rate has never been clearly understood, so Marra and colleagues set out to compare the genomes between these ancient living fish and try to determine a genomic basis.
Three major differences in the immune systems between elasmobranches, teleosts and higher order vertebrates are:
- A lack of bone marrow in elasmobranches (where B cells normally develop)
- The presence of a newly discovered form of immunoglobulin (IgNAR) which lacks light chain structures
- Major differences in the genomic organization of immunology genes
Another adaptive change that is present in ancient fish concerns their cardiovascular system, and the evolutionary development of a system designed to preserve regional endothermy in an icy ocean. This was the underlying reason the researchers chose to use tissue from the heart muscle for comparison; so they could use the heart transcriptome to examine a large repertoire of genes for possible evidence of convergent evolution between each of the fish species.

On a genomic level, the researchers more specific goals were exploring differences in expressed gene content between elasmobranchs and teleosts, finding differences between specific species in regards to particular phenomena of interest such as adaptive immunity and wound healing, and finally identify genes with a history of molecular adaptation.
Once heart tissue was collected, sequencing libraries were constructed and compared via paired-end sequencing. The sequencing was pooled with four species each, and then separated per species to create a species-specific heart transcriptome. The genes were run through BLAST and the results were analyzed and compared though genomic databases to compare and cluster.
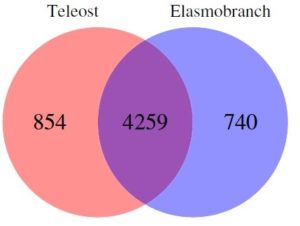
Marra and colleagues discovered that while there is significant overlap, there are clear genetic differences between these species, particularly elasmobraches and teleosts. Furthermore, elasmobraches contain exclusive genes involved in immunity, metabolism and gene regulation within the heart genome, which make up a substantial proportion of the genes expressed in that tissue.
The researchers also revealed that while each of the species shared approximately 50% of their genomes in heart tissue, 50% were different. It is of particular note the immune genes in particular showed the most difference between classes of fish, both at the genomic and transcription levels.
While the researchers did not find IgNAR using the Swiss-plot databases, they did find IgW (a cartilaginous fish version of the IgD found in mammals), and legumain (LGMN), a gene related to antigen presentation in MHC class II molecules which has also been reported to be overexpressed in several cancers. It is suspected that IgNAR was not found due to it not being recognized by the database.
Legumain is also used as a target to eliminate tumor associated macrophages, and there are several studies reporting various shark extracts as having anti-tumorigenic properties. Overexpression of legumain occurs in tumor-associated macrophages and is thought to contribute to the tumor promoting inflammation associated with most malignancies.
Other genes of interest included oncogene Bag1, an athanogene which prevents cell apoptosis; Tim22, which regulates mitochondrial control; and a group of genes which regulate damaging cell peroxidases. It is suspected that Bag1 has evolved within the shark genome to be better regulated via highly controlled expression levels so that the cell cycle can be closely monitored, unlike more advanced vertebrates where Bag1 overexpression can cause cancer development. In regards to wound healing, there are several pathways which must act in tandem in order for this system to function properly, including inflammation and pathogen clearing. In particular, elasmobrach MHC class II molecules (used for immune cell signaling), have evolved to be more specific and tightly controlled then in higher level vertebrates. This MHC class II specialization is suspected to enhance wound repair.
The collection of highly regulatory genes found exclusively in ancient fish species showcases how little scientists know about the earliest vertebrates, and how much information can be translated into human cellular repair. While researchers have made substantial progress on understanding cancer pathways, they have not yet made leeway on methods to reverse cellular damage and initiate repair, making the genome of elasmobranches an invaluable resource in cancer treatment and information.
Comments